HydPy-G-GR4 (Génie Rural model with 4 parameters)¶
The GR4J model (modèle du Génie Rural à 4 parametres Journalier) is a daily
lumped four-parameter rainfall-runoff model and belongs to the family of soil moisture
accounting models. It was initially published by Perrin et al. (2003) and is a
modification of GR3J. Our implementation, gland_gr4, follows the one of the R
package airGR (Coron et al., 2017) but with a few extensions. Namely, it applies to
arbitrary simulation step sizes and offers more flexibility in simulating interception
and runoff concentration.
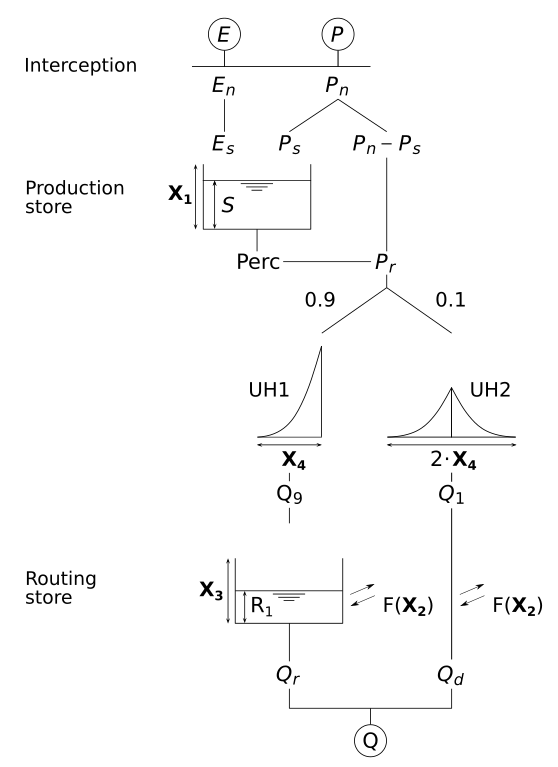
The following figure (Coron et al., 2023) shows the general structure of
gland_gr4:
Integration tests¶
Note
When new to HydPy, consider reading section Integration Tests first.
The following integration tests rely on the example dataset L0123001 of airGR and allow for comparing both model implementations.
All tests cover 50 days with a simulation step of one day:
>>> from hydpy import pub
>>> pub.timegrids = "1990-01-01", "1990-02-20", "1d"
Prepare a model instance and pass it to an element connected to a single outlet node:
>>> from hydpy.models.gland_gr4 import *
>>> parameterstep("1d")
>>> from hydpy import Element
>>> land = Element("land", outlets="outlet")
>>> land.model = model
The size of the (lumpy simulated) catchment is 360 km²:
>>> area(360.0)
gland_gr4 requires a submodel that provides potential evaporation estimates. We
select evap_ret_io, which allows using a predefined time series:
>>> with model.add_petmodel_v1("evap_ret_io"):
... evapotranspirationfactor(1.0)
Initialise a test function object, which prepares and runs the tests and prints their results for the selected sequences:
>>> from hydpy import IntegrationTest
>>> test = IntegrationTest(land)
>>> test.plotting_options.axis1 = inputs.p, fluxes.e, fluxes.qh
>>> test.dateformat = "%d.%m."
The mentioned precipitation and potential evapotranspiration time series stem from the mentioned dataset:
>>> inputs.p.series = (
... 0.0, 9.3, 3.2, 7.3, 0.0, 0.0, 0.0, 0.0, 0.1, 0.2, 2.9, 0.2, 0.0, 0.0, 0.0, 3.3,
... 4.6, 0.8, 1.8, 1.1, 0.0, 5.0, 13.1, 14.6, 4.0, 0.8, 0.1, 3.3, 7.7, 10.3, 3.7,
... 15.3, 3.2, 2.7, 2.2, 8.0, 14.3, 6.3, 0.0, 5.9, 9.2, 6.1, 0.1, 0.0, 2.8, 10.6,
... 8.8, 7.2, 4.9, 1.8
... )
>>> model.petmodel.sequences.inputs.referenceevapotranspiration.series = (
... 0.3, 0.4, 0.4, 0.3, 0.1, 0.1, 0.1, 0.2, 0.2, 0.3, 0.3, 0.2, 0.2, 0.2, 0.2, 0.3,
... 0.3, 0.2, 0.2, 0.3, 0.2, 0.2, 0.3, 0.6, 0.4, 0.4, 0.4, 0.5, 0.4, 0.3, 0.3, 0.5,
... 0.5, 0.3, 0.3, 0.4, 0.4, 0.3, 0.2, 0.1, 0.1, 0.0, 0.1, 0.1, 0.0, 0.2, 0.9, 0.9,
... 0.5, 0.9
... )
base example¶
We take the following values of the first three of four original GR4J parameters X1,
X2, and X3 directly from the airGR manual:
>>> x1(257.238)
>>> x2(1.012)
>>> x3(88.235)
The parameter IMax is unavailable in GR4J, which always assumes an interception
storage of zero capacity. Hence, consistency with GR4J requires setting IMax to
zero:
>>> imax(0.0)
The fourth original GR4J parameter, X4, defines the time base of two Unit
Hydrographs. Unlike GR4J, gland_gr4 does not rely on hard-coded Unit Hydrographs and
has no parameter named X4 but provides two optional submodel ports that allow adding
different types of runoff concentration submodels. For consistency with GR4J, you can
select the rconc_uh submodel for both ports. Its options gr_uh2 and gr_uh1
correspond to the typical configuration of the Unit Hydrographs of the direct flow and
the routing store component, respectively. Passing the value of the original X4
parameter as a function argument in both cases completes the desired consistency (note
that, for non-daily simulations, you might also want to adjust the other function
argument beta, which defaults to 2.5):
>>> with model.add_rconcmodel_directflow_v1("rconc_uh"):
... uh("gr_uh2", x4=2.208)
>>> with model.add_rconcmodel_routingstore_v1("rconc_uh"):
... uh("gr_uh1", x4=2.208)
Now that all models have been selected and parameterised, we can define their initial conditions:
>>> test.inits = (
... (states.i, 0.0),
... (states.s, 0.3 * x1),
... (states.r, 0.5 * x3),
... (model.rconcmodel_routingstore.sequences.logs.quh, 0.0),
... (model.rconcmodel_directflow.sequences.logs.quh, 0.0),
... )
The following simulation results agree with those of the GR4J airGR implementation:
>>> conditions = test("gland_gr4_base_example", get_conditions="1990-01-01")
Click to see the table
Click to see the graphThere is no indication of an error in the water balance:
>>> from hydpy import round_
>>> round_(model.check_waterbalance(conditions))
0.0
groundwater loss¶
In the base example, the groundwater exchange coefficient X2 is
positive, which results in a groundwater gain. Here, we demonstrate the opposite case
of a net groundwater loss and start with empty production and routing stores:
>>> x2(-1.012)
>>> test.inits.s = 0.0
>>> test.inits.r = 0.0
>>> conditions = test("gland_gr4_groundwater_loss", get_conditions="1990-01-01")
Click to see the table
Click to see the graphThere is no indication of an error in the water balance:
>>> round_(model.check_waterbalance(conditions))
0.0
interception¶
This integration test repeats the base example with a non-zero interception capacity:
>>> imax(10.0)
>>> x2(1.012)
>>> test.inits.s = 0.3 * x1
>>> test.inits.r = 0.5 * x3
>>> conditions = test("gland_gr4_interception", get_conditions="1990-01-01")
Click to see the table
Click to see the graphThere is no indication of an error in the water balance:
>>> round_(model.check_waterbalance(conditions))
0.0
no rconc submodels¶
As mentioned above, the two runoff concentration ports are optional, meaning that
gland_gr4 can work without the corresponding submodels. In this case, it neglects
any related runoff concentration, resulting in an earlier outflow peak then in the last
example:
>>> model.rconcmodel_directflow = None
>>> model.rconcmodel_routingstore = None
>>> conditions = test("gland_gr4_no_rconc_submodels", get_conditions="1990-01-01")
Click to see the table
Click to see the graphThere is no indication of an error in the water balance:
>>> round_(model.check_waterbalance(conditions))
0.0
- class hydpy.models.gland_gr4.Model[source]¶
Bases:
Main_PETModel_V1,Main_RConcModel_V2HydPy-G-GR4 (Génie Rural model with 4 parameters).
- The following “run methods” are called in the given sequence during each simulation step:
Calc_E_V1Let a submodel that conforms to thePETModel_V1interface calculate the potential evapotranspiration.Calc_EI_V1Calculate the actual evaporation from the interception store.Calc_PN_V1Calculate the net precipitation by considering all interception losses.Calc_EN_V1Calculate the net evapotranspiration capacity by considering interception evaporation.Update_I_V1Update the interception store based on precipitation, net precipitation, and interception evaporation.Calc_PS_V1Calculate the part of net precipitation filling the production store.Calc_ES_V1Calculate the actual evapotranspiration from the production store.Update_S_V1Update the production store by adding precipitation and evapotranspiration.Calc_Perc_V1Calculate the percolation from the production store.Update_S_V2Update the production store by subtracting percolation.Calc_AE_V1Calculate the total actual evapotranspiration.Calc_Pr_V1Calculate the total inflow into the runoff concentration module.Calc_PR1_PR9_V1SplitPRintoPR1andPR9.Calc_Q9_V1TransformPR9intoQ9.Calc_Q1_V1TransformPR1intoQ1.Calc_FR_V1Calculate the groundwater exchange affecting the routing store according to GR4.Update_R_V1Update the level of the non-linear routing store by adding its inflows according to GR4 and GR5.Calc_QR_V1Calculate the outflow of the routing store.Update_R_V3Update the non-linear routing store by subtracting its outflow.Calc_FD_V1Calculate the groundwater exchange affecting the direct runoff.Calc_QD_V1Calculate the direct runoff.Calc_QH_V1Calculate the total runoff according to GR4 and GR5.Calc_QV_V1Calculate total discharge in m³/s.
- The following “outlet update methods” are called in the given sequence at the end of each simulation step:
Pass_Q_V1Update the outlet link sequence.
- The following “additional methods” might be called by one or more of the other methods or are meant to be directly called by the user:
Calc_E_PETModel_V1Let a submodel that conforms to thePETModel_V1interface calculate the potential evapotranspiration.Calc_Q_RConcModel_V1Let a submodel that follows theRConcModel_V1submodel interface perform runoff concentration.
- Users can hook submodels into the defined main model if they satisfy one of the following interfaces:
PETModel_V1Simple interface for calculating all potential evapotranspiration values in one step.RConcModel_V1Simple interface for calculating runoff concentration processes.
- DOCNAME = ('G-GR4', 'Génie Rural model with 4 parameters')¶
- petmodel¶
Required submodel that complies with the following interface: PETModel_V1.
- rconcmodel_routingstore¶
Optional submodel that complies with the following interface: RConcModel_V1.
- rconcmodel_directflow¶
Optional submodel that complies with the following interface: RConcModel_V1.
- check_waterbalance(initial_conditions: dict[str, dict[str, dict[str, float | ndarray[tuple[Any, ...], dtype[float64]]]]]) float[source]¶
Determine the water balance error of the previous simulation run in mm.
Method
check_waterbalance()calculates the balance error as follows:\[\begin{split}Error = \Sigma InOut - \Delta Vol - \Delta Rconc \\ \\ \Sigma InOut = \sum_{t=t0}^{t1} P_t - AE_t + FR_t + FD_t - QH_t \\ \Delta Vol = \left( I_{t1} - I_{t0} \right) + \left( S_{t1} - S_{t0} \right) + \left( R_{t1} - R_{t0} \right) \\ \Delta Rconc = get\_waterbalance_{direct \ flow}(*) + get\_waterbalance_{routing \ store}(*)\end{split}\]
- REUSABLE_METHODS = ()¶
- class hydpy.models.gland_gr4.ControlParameters(master: Parameters, cls_fastaccess: type[FastAccessParameter] | None = None, cymodel: CyModelProtocol | None = None)¶
Bases:
SubParametersControl parameters of model gland_gr4.
- The following classes are selected:
- class hydpy.models.gland_gr4.DerivedParameters(master: Parameters, cls_fastaccess: type[FastAccessParameter] | None = None, cymodel: CyModelProtocol | None = None)¶
Bases:
SubParametersDerived parameters of model gland_gr4.
- class hydpy.models.gland_gr4.FluxSequences(master: Sequences, cls_fastaccess: type[TypeFastAccess_co] | None = None, cymodel: CyModelProtocol | None = None)¶
Bases:
FluxSequencesFlux sequences of model gland_gr4.
- The following classes are selected:
E()Potential evapotranspiration [mm/T].EN()Net evapotranspiration capacity [mm/T].PN()Net precipitation [mm/T].EI()Actual evaporation from the interception store [mm/T].ES()Actual evapotranspiration from the production store [mm/T].AE()Total actual evapotranspiration [mm/T].PR()Total inflow into the runoff concentration module [mm/T].Perc()Percolation [mm/T].Q9()Outflow of runoff concentration submodel receivingPR9[mm/T].Q1()Outflow of runoff concentration submodel receivingPR1[mm/T].FD()Groundwater exchange affecting the direct runoff [mm/T].FR()Groundwater exchange affecting the routing store [mm/T].QR()Outflow of the routing store [mm/T].QD()Direct runoff [mm/T].QH()Total runoff [mm/T].QV()Total discharge [m³/s].
- class hydpy.models.gland_gr4.InputSequences(master: Sequences, cls_fastaccess: type[TypeFastAccess_co] | None = None, cymodel: CyModelProtocol | None = None)¶
Bases:
InputSequencesInput sequences of model gland_gr4.
- The following classes are selected:
P()Precipitation [mm/T].
- class hydpy.models.gland_gr4.OutletSequences(master: Sequences, cls_fastaccess: type[TypeFastAccess_co] | None = None, cymodel: CyModelProtocol | None = None)¶
Bases:
OutletSequencesOutlet sequences of model gland_gr4.
- The following classes are selected:
Q()Runoff [m³/s].
- class hydpy.models.gland_gr4.StateSequences(master: Sequences, cls_fastaccess: type[TypeFastAccess_co] | None = None, cymodel: CyModelProtocol | None = None)¶
Bases:
StateSequencesState sequences of model gland_gr4.
