devicetools¶
This modules implements the fundamental features for structuring HydPy projects.
Module devicetools provides two Device subclasses, Node and Element. In this
documentation, “node” stands for an object of class Node, “element” for an object of
class Element, and “device” for either of them (you cannot initialise objects of
class Device directly). On the other hand, the term “nodes”, for example, does not
necessarily mean an object of class Nodes but any other group of Node objects as
well.
Each element handles a single Model object and represents, for example, a subbasin or
a channel segment. The purpose of a node is to connect different elements and, for
example, to pass the discharge calculated for a subbasin outlet (from a first element)
to the top of a channel segment (to second element). Class Node and Element come
with specialised container classes (Nodes and Elements). The names of individual
nodes and elements serve as identity values, so duplicate names are not permitted.
Note that module devicetools implements a registry mechanism both for nodes and
elements, preventing instantiating an object with an already assigned name. This
mechanism allows to address the same node or element in different network files (see
module selectiontools).
Let us take class Node as an example. One can call its constructor with the same
name multiple times, but it returns already existing nodes when available:
>>> from hydpy import Node
>>> node1 = Node("test1")
>>> node2a = Node("test2")
>>> node2b = Node("test2")
>>> node1 is node2a
False
>>> node2a is node2b
True
To get information on all currently registered nodes, call method
extract_new():
>>> Node.extract_new()
Nodes("test1", "test2")
Method extract_new() returns only those nodes prepared or
recovered after its last invocation:
>>> node1 = Node("test1")
>>> node3a = Node("test3")
>>> Node.extract_new()
Nodes("test1", "test3")
For a complete list of all available nodes, use the method query_all():
>>> Node.query_all()
Nodes("test1", "test2", "test3")
When working interactively in the Python interpreter, it might sometimes be helpful to clear the registry entirely. However, Do this with care because defining nodes with already assigned names might result in surprises due to using their names for identification:
>>> nodes = Node.query_all()
>>> Node.clear_all()
>>> Node.query_all()
Nodes()
>>> node3b = Node("test3")
>>> node3b in nodes
True
>>> nodes.test3.name == node3b.name
True
>>> nodes.test3 is node3b
False
Module devicetools implements the following members:
TypeDeviceType variable.
TypeDevicesType variable.
TypeNodeElementType variable.
KeywordsSet of keyword arguments used to describe and search forElementandNodeobjects.
FusedVariableCombinesInputSequence,ReceiverSequence, andOutputSequencesubclasses of different models dealing with the same property in a single variable.
clear_registries_temporarily()Context manager for clearing the currentNode,Element, andFusedVariableregistries.
ElementHandles aModelobject and connects it to other models viaNodeobjects.
- class hydpy.core.devicetools.Keywords(*names: str)[source]¶
-
Set of keyword arguments used to describe and search for
ElementandNodeobjects.- startswith(name: str) list[str][source]¶
Return a list of all keywords, starting with the given string.
>>> from hydpy.core.devicetools import Keywords >>> keywords = Keywords("first_keyword", "second_keyword", ... "keyword_3", "keyword_4", ... "keyboard") >>> keywords.startswith("keyword") ['keyword_3', 'keyword_4']
- endswith(name: str) list[str][source]¶
Return a list of all keywords ending with the given string.
>>> from hydpy.core.devicetools import Keywords >>> keywords = Keywords("first_keyword", "second_keyword", ... "keyword_3", "keyword_4", ... "keyboard") >>> keywords.endswith("keyword") ['first_keyword', 'second_keyword']
- contains(name: str) list[str][source]¶
Return a list of all keywords containing the given string.
>>> from hydpy.core.devicetools import Keywords >>> keywords = Keywords("first_keyword", "second_keyword", ... "keyword_3", "keyword_4", ... "keyboard") >>> keywords.contains("keyword") ['first_keyword', 'keyword_3', 'keyword_4', 'second_keyword']
- update(*names: str) None[source]¶
Before updating, the given names are checked to be valid variable identifiers.
>>> from hydpy.core.devicetools import Keywords >>> keywords = Keywords("first_keyword", "second_keyword", ... "keyword_3", "keyword_4", ... "keyboard") >>> keywords.update("test_1", "test 2") Traceback (most recent call last): ... ValueError: While trying to add the keyword `test 2` to device ?, the following error occurred: The given name string `test 2` does not define a valid variable identifier. ...
Note that even the first string (test1) is not added due to the second one (test 2) being invalid.
>>> keywords Keywords("first_keyword", "keyboard", "keyword_3", "keyword_4", "second_keyword")
After correcting the second string, everything works fine:
>>> keywords.update("test_1", "test_2") >>> keywords Keywords("first_keyword", "keyboard", "keyword_3", "keyword_4", "second_keyword", "test_1", "test_2")
- add(name: Any) None[source]¶
Before adding a new name, it is checked to be a valid variable identifier.
>>> from hydpy.core.devicetools import Keywords >>> keywords = Keywords("first_keyword", "second_keyword", ... "keyword_3", "keyword_4", ... "keyboard") >>> keywords.add("1_test") Traceback (most recent call last): ... ValueError: While trying to add the keyword `1_test` to device ?, the following error occurred: The given name string `1_test` does not define a valid variable identifier. ...
>>> keywords Keywords("first_keyword", "keyboard", "keyword_3", "keyword_4", "second_keyword")
After correcting the string, everything works fine:
>>> keywords.add("one_test") >>> keywords Keywords("first_keyword", "keyboard", "keyword_3", "keyword_4", "one_test", "second_keyword")
- class hydpy.core.devicetools.FusedVariable(name: str, *sequences: sequencetools.InOutSequenceTypes)[source]¶
Bases:
objectCombines
InputSequence,ReceiverSequence, andOutputSequencesubclasses of different models dealing with the same property in a single variable.Class
FusedVariableis one possible type of propertyvariableof classNode. We need it in some HydPy projects where the involved models not only pass runoff to each other but also share other types of data. Each project-specificFusedVariableobject serves as a “meta-type”, indicating which input and output sequences of the different models correlate and are thus connectable.Using class
FusedVariableis easiest to explain by a concrete example. Assume we useconv_nnto interpolate the air temperature for a specific location. We use this temperature as input to anmeteo_temp_iomodel, which passes it to anevap_ret_fao56model, which requires this and other meteorological data to calculate potential evapotranspiration. Further, we pass the estimated potential evapotranspiration as input tolland_ddfor calculating the actual evapotranspiration, which receives it through a submodel instance ofevap_ret_io. Hence, we need to connect the output sequenceMeanReferenceEvapotranspirationofevap_ret_fao56with the input sequenceReferenceEvapotranspirationofevap_ret_io.- ToDo: This example needs to be updated. Today one could directly use
evap_ret_fao56as a submodel oflland_dd. However, it still demonstrates the relevant connection mechanisms correctly.
Additionally,
lland_ddrequires temperature data itself for modelling snow processes, introducing the problem that we need to use the same data (the output ofconv_nn) as the input of two differently named input sequences (TemperatureandTemLformeteo_temp_ioandlland_dd, respectively).We need to create two
FusedVariableobjects, for our concrete example. E combinesMeanReferenceEvapotranspirationandReferenceEvapotranspirationand T combinesTemperatureandTemL(for convenience, we import their globally available aliases):>>> from hydpy import FusedVariable >>> from hydpy.aliases import ( ... evap_inputs_ReferenceEvapotranspiration, meteo_inputs_Temperature, ... lland_inputs_TemL, evap_fluxes_MeanReferenceEvapotranspiration) >>> E = FusedVariable("E", evap_inputs_ReferenceEvapotranspiration, ... evap_fluxes_MeanReferenceEvapotranspiration) >>> T = FusedVariable("T", meteo_inputs_Temperature, lland_inputs_TemL)
Now we can construct the network:
Node t1 handles the original temperature data and serves as the input node to element conv. We define the (arbitrarily selected) string Temp to be its variable.
Node e receives the potential evapotranspiration calculated by element evap and passes it to element lland. Node e thus receives the fused variable E.
Node t2 handles the interpolated temperature and serves as the outlet node of element conv and the input node to elements evap and lland. Node t2 thus receives the fused variable T.
>>> from hydpy import Node, Element >>> t1 = Node("t1", variable="Temp") >>> t2 = Node("t2", variable=T) >>> e = Node("e", variable=E) >>> conv = Element("element_conv", inlets=t1, outlets=t2) >>> evap = Element("element_evap", inputs=t2, outputs=e) >>> lland = Element("element_lland", inputs=(t2, e), outlets="node_q")
Now we can prepare the different model objects and assign them to their corresponding elements (note that parameters
InputCoordinatesandOutputCoordinatesofconv_nnfirst require information on the location of the relevant nodes):>>> from hydpy import prepare_model >>> model_conv = prepare_model("conv_nn") >>> model_conv.parameters.control.inputcoordinates(t1=(0, 0)) >>> model_conv.parameters.control.outputcoordinates(t2=(1, 1)) >>> model_conv.parameters.control.maxnmbinputs(1) >>> model_conv.parameters.update() >>> conv.model = model_conv >>> model = prepare_model("evap_ret_fao56") >>> model.tempmodel = prepare_model("meteo_temp_io") >>> evap.model = model >>> model = prepare_model("lland_dd") >>> model.aetmodel = prepare_model("evap_aet_minhas") >>> model.aetmodel.petmodel = prepare_model("evap_ret_io") >>> lland.model = model
We assign a temperature value to node t1:
>>> t1.sequences.sim = -273.15
Model
conv_nncan now perform a simulation step and pass its output to node t2:>>> conv.model.simulate(0) >>> t2.sequences.sim sim(-273.15)
Without further configuration,
evap_ret_fao56cannot perform any simulation steps. Hence, we just call itsload_data()method to show that the input sequenceTemperatureof its submodel is well-connected to theSimsequence of node t2 and receives the correct data:>>> evap.model.load_data(0) >>> evap.model.tempmodel.sequences.inputs.temperature temperature(-273.15)
The output sequence
MeanReferenceEvapotranspirationis also well connected. A call to methodupdate_outputs()passes its (manually set) value to node e, respectively:>>> evap.model.sequences.fluxes.meanreferenceevapotranspiration = 999.9 >>> evap.model.update_outputs() >>> e.sequences.sim sim(999.9)
Finally, both input sequences
TemLandReferenceEvapotranspirationreceive the current values of nodes t2 and e:>>> lland.model.load_data(0) >>> lland.model.sequences.inputs.teml teml(-273.15) >>> lland.model.aetmodel.petmodel.sequences.inputs.referenceevapotranspiration referenceevapotranspiration(999.9)
When defining fused variables, class
FusedVariableperforms some registration behind the scenes, similar to what classesNodeandElementdo. Again, the name works as the identifier, and we force the same fused variable to exist only once, even when defined in different selection files repeatedly. Hence, when we repeat the definition from above, we get the same object:>>> Test = FusedVariable("T", meteo_inputs_Temperature, lland_inputs_TemL) >>> T is Test True
Changing the member sequences of an existing fused variable is not allowed:
>>> from hydpy.aliases import hland_inputs_T >>> FusedVariable("T", hland_inputs_T, lland_inputs_TemL) Traceback (most recent call last): ... ValueError: The sequences combined by a FusedVariable object cannot be changed. The already defined sequences of the fused variable `T` are `lland_inputs_TemL and meteo_inputs_Temperature` instead of `hland_inputs_T and lland_inputs_TemL`. Keep in mind, that `name` is the unique identifier for fused variable instances.
Defining additional fused variables with the same member sequences is not advisable, but is allowed:
>>> Temp = FusedVariable("Temp", meteo_inputs_Temperature, lland_inputs_TemL) >>> T is Temp False
To get an overview of the existing fused variables, call method
get_registry():>>> len(FusedVariable.get_registry()) 3
Principally, you can clear the registry via method
clear_registry(), but remember it does not removeFusedVariableobjects from the running process being otherwise referenced:>>> FusedVariable.clear_registry() >>> FusedVariable.get_registry() () >>> t2.variable FusedVariable("T", lland_inputs_TemL, meteo_inputs_Temperature)
For convenience,
FusedVariablesupports equality comparisons with other fused variables (Are all handled variable types identical?), individual sequences (Is the given sequence type among the handled types?), and sequence names (Is any of the handled types the given name?):>>> assert T == Temp >>> assert T != E >>> assert T == meteo_inputs_Temperature >>> assert meteo_inputs_Temperature == T >>> assert T != evap_inputs_ReferenceEvapotranspiration >>> assert evap_inputs_ReferenceEvapotranspiration != T >>> assert T == "TemL" >>> assert "TemL" == T >>> assert T != "ReferenceEvapotranspiration" >>> assert "ReferenceEvapotranspiration" != T
- classmethod get_registry() tuple[FusedVariable, ...][source]¶
Get all
FusedVariableobjects initialised so far.
- classmethod clear_registry() None[source]¶
Clear the registry from all
FusedVariableobjects initialised so far.Use this method only for good reasons!
- class hydpy.core.devicetools.Devices(*values: TypeDevice | str | Iterable[TypeDevice | str] | None, mutable: bool = True)[source]¶
Bases:
Generic[TypeDevice]Base class for class
Elementsand classNodes.The following features are common to class
Nodesand classElements. We arbitrarily select classNodesfor all examples.To initialise a
Nodescollection, pass a variable number ofstrorNodeobjects. Strings are used to create new or query already existing nodes automatically:>>> from hydpy import Node, Nodes >>> nodes = Nodes("na", ... Node("nb", variable="W"), ... Node("nc", keywords=("group_a", "group_1")), ... Node("nd", keywords=("group_a", "group_2")), ... Node("ne", keywords=("group_b", "group_1")))
Nodesinstances are containers supporting attribute and item access. You can access each node directly by its name:>>> nodes.na Node("na", variable="Q") >>> nodes["na"] Node("na", variable="Q")
In many situations, a
Nodesinstance contains a single node only. One can query such a single node using zero as the index for convenience:>>> Nodes("na")[0] Node("na", variable="Q")
Other number-based indexed are not allowed:
>>> Nodes("na", "nb")[1] Traceback (most recent call last): ... KeyError: 'Indexing with other numbers than `0` is not supported but `1` is given.'
An automatic check prevents unexpected results when applying zero-based indexing on
Nodesinstances containing multiple nodes:>>> Nodes("na", "nb")[0] Traceback (most recent call last): ... KeyError: 'Indexing with `0` is only safe for Node handlers containing a single Node.'
Wrong node names result in the following error messages:
>>> nodes.wrong Traceback (most recent call last): ... AttributeError: The selected Nodes object has neither a `wrong` attribute nor does it handle a Node object with name or keyword `wrong`, which could be returned. >>> nodes["wrong"] Traceback (most recent call last): ... KeyError: 'No node named `wrong` available.'
As explained in more detail in the documentation on property
keywords, you can also use the keywords of the individual nodes to query the relevant ones:>>> nodes.group_a Nodes("nc", "nd")
You can remove nodes both via the attribute and item syntax:
>>> "na" in nodes True >>> del nodes.na >>> "na" in nodes False >>> del nodes.na Traceback (most recent call last): ... AttributeError: The actual Nodes object does not handle a Node object named `na` which could be removed, and deleting other attributes is not supported.
>>> nodes.add_device("na") >>> del nodes["na"] >>> del nodes["na"] Traceback (most recent call last): ... KeyError: 'No node named `na` available.'
However, as shown by the following example, setting devices via attribute assignment or item assignment could result in inconsistencies and is thus not allowed (see method
add_device()instead):>>> nodes.NF = Node("nf") Traceback (most recent call last): ... AttributeError: Setting attributes of Nodes objects could result in confusion whether a new attribute should be handled as a Node object or as a "normal" attribute and is thus not support, hence `NF` is rejected. >>> nodes["NF"] = Node("nf") Traceback (most recent call last): ... TypeError: 'Nodes' object does not support item assignment
Nodesinstances support iteration:>>> len(nodes) 4 >>> for node in nodes: ... print(node.name, end=",") nb,nc,nd,ne,
The binary operators +, +=, -, and -= support adding and removing single devices or groups of devices:
>>> nodes Nodes("nb", "nc", "nd", "ne") >>> nodes - Node("nc") Nodes("nb", "nd", "ne")
Nodes(“nb”, “nc”, “nd”, “ne”) >>> nodes -= Nodes(“nc”, “ne”) >>> nodes Nodes(“nb”, “nd”)
>>> nodes + "nc" Nodes("nb", "nc", "nd") >>> nodes Nodes("nb", "nd") >>> nodes += ("nc", Node("ne")) >>> nodes Nodes("nb", "nc", "nd", "ne")
Attempts to add already existing or to remove non-existing devices do no harm:
>>> nodes Nodes("nb", "nc", "nd", "ne") >>> nodes + ("nc", "ne") Nodes("nb", "nc", "nd", "ne") >>> nodes - Node("na") Nodes("nb", "nc", "nd", "ne")
Comparisons are supported, with “x < y” being
Trueif “x” is a subset of “y”:>>> subgroup = Nodes("nc", "ne") >>> subgroup < nodes, nodes < subgroup, nodes < nodes (True, False, False) >>> subgroup <= nodes, nodes <= subgroup, nodes <= nodes (True, False, True) >>> subgroup == nodes, nodes == subgroup, nodes == nodes, nodes == "nodes" (False, False, True, False) >>> subgroup != nodes, nodes != subgroup, nodes != nodes, nodes != "nodes" (True, True, False, True) >>> subgroup >= nodes, nodes >= subgroup, nodes >= nodes (False, True, True) >>> subgroup > nodes, nodes > subgroup, nodes > nodes (False, True, False)
Class
Nodessupports the in operator both forstrandNodeobjects and generally returnsFalsefor other types:>>> "na" in nodes False >>> "nb" in nodes True >>> Node("na") in nodes False >>> Node("nb") in nodes True >>> 1 in nodes False
Passing wrong arguments to the constructor of class
Noderesults in errors like the following:>>> from hydpy import Element >>> Nodes("na", Element("ea")) Traceback (most recent call last): ... TypeError: While trying to initialise a `Nodes` object, the following error occurred: The given (sub)value `Element("ea")` is not an instance of the following classes: Node and str.
- add_device(device: TypeDevice | str, force: bool = False) None[source]¶
Add the given
NodeorElementobject to the actualNodesorElementsobject.You can pass either a string or a device:
>>> from hydpy import Nodes >>> nodes = Nodes() >>> nodes.add_device("old_node") >>> nodes Nodes("old_node") >>> nodes.add_device("new_node") >>> nodes Nodes("new_node", "old_node")
Method
add_device()is disabled for immutableNodesandElementsobjects by default:>>> nodes._mutable = False >>> nodes.add_device("newest_node") Traceback (most recent call last): ... RuntimeError: While trying to add the device `newest_node` to a Nodes object, the following error occurred: Adding devices to immutable Nodes objects is not allowed.
Use parameter force to override this safety mechanism if necessary:
>>> nodes.add_device("newest_node", force=True) >>> nodes Nodes("new_node", "newest_node", "old_node")
- remove_device(device: TypeDevice | str, force: bool = False) None[source]¶
Remove the given
NodeorElementobject from the actualNodesorElementsobject.You can pass either a string or a device:
>>> from hydpy import Node, Nodes >>> nodes = Nodes("node_x", "node_y") >>> node_x, node_y = nodes >>> nodes.remove_device(Node("node_y")) >>> nodes Nodes("node_x") >>> nodes.remove_device(Node("node_x")) >>> nodes Nodes() >>> nodes.remove_device(Node("node_z")) Traceback (most recent call last): ... ValueError: While trying to remove the device `node_z` from a Nodes object, the following error occurred: The actual Nodes object does not handle such a device.
Method
remove_device()is disabled for immutableNodesandElementsobjects by default:>>> nodes.add_device(node_x) >>> nodes._mutable = False >>> nodes.remove_device("node_x") Traceback (most recent call last): ... RuntimeError: While trying to remove the device `node_x` from a Nodes object, the following error occurred: Removing devices from immutable Nodes objects is not allowed. >>> nodes Nodes("node_x")
Use parameter force to override this safety mechanism if necessary:
>>> nodes.remove_device("node_x", force=True) >>> nodes Nodes()
- property names: tuple[str, ...]¶
A sorted tuple of the names of the handled devices.
>>> from hydpy import Nodes >>> Nodes("a", "c", "b").names ('a', 'b', 'c')
- property devices: tuple[TypeDevice, ...]¶
A tuple of the handled devices sorted by the device names.
>>> from hydpy import Nodes >>> for node in Nodes("a", "c", "b").devices: ... print(repr(node)) Node("a", variable="Q") Node("b", variable="Q") Node("c", variable="Q")
- property keywords: set[str]¶
A set of all keywords of all handled devices.
In addition to attribute access via device names,
NodesandElementsobjects allow for attribute access via keywords, allowing for an efficient search of certain groups of devices. Let us use the example from above, where the nodes na and nb have no keywords, but each of the other three nodes both belongs to either group_a or group_b and group_1 or group_2:>>> from hydpy import Node, Nodes >>> nodes = Nodes("na", ... Node("nb", variable="W"), ... Node("nc", keywords=("group_a", "group_1")), ... Node("nd", keywords=("group_a", "group_2")), ... Node("ne", keywords=("group_b", "group_1"))) >>> nodes Nodes("na", "nb", "nc", "nd", "ne") >>> sorted(nodes.keywords) ['group_1', 'group_2', 'group_a', 'group_b']
If you are interested in inspecting all devices belonging to group_a, select them via this keyword:
>>> subgroup = nodes.group_1 >>> subgroup Nodes("nc", "ne")
You can further restrict the search by also selecting the devices belonging to group_b, which holds only for node “e”, in the discussed example:
>>> subsubgroup = subgroup.group_b >>> subsubgroup Node("ne", variable="Q", keywords=["group_1", "group_b"])
Note that the keywords already used for building a device subgroup are not informative anymore (as they hold for each device) and are thus not shown anymore:
>>> sorted(subgroup.keywords) ['group_a', 'group_b']
The latter might be confusing if you intend to work with a device subgroup for a longer time. After copying the subgroup, all keywords of the contained devices are available again:
>>> from copy import copy >>> newgroup = copy(subgroup) >>> sorted(newgroup.keywords) ['group_1', 'group_a', 'group_b']
- search_keywords(*keywords: str) TypeDevices[source]¶
Search for all devices handling at least one of the given keywords and return them.
>>> from hydpy import Node, Nodes >>> nodes = Nodes("na", ... Node("nb", variable="W"), ... Node("nc", keywords=("group_a", "group_1")), ... Node("nd", keywords=("group_a", "group_2")), ... Node("ne", keywords=("group_b", "group_1"))) >>> nodes.search_keywords("group_c") Nodes() >>> nodes.search_keywords("group_a") Nodes("nc", "nd") >>> nodes.search_keywords("group_a", "group_1") Nodes("nc", "nd", "ne")
- copy() TypeDevices[source]¶
Return a shallow copy of the actual
NodesorElementsobject.Method
copy()returns a semi-flat copy ofNodesorElementsobjects due to their devices being not copyable:>>> from hydpy import Nodes >>> old = Nodes("x", "y") >>> import copy >>> new = copy.copy(old) >>> new == old True >>> new is old False >>> new.devices is old.devices False >>> new.x is new.x True
Changing the
nameof a device is recognised both by the original and the copied collection objects:>>> new.x.name = "z" >>> old.z Node("z", variable="Q") >>> new.z Node("z", variable="Q")
Deep copying is permitted due to the above reason:
>>> copy.deepcopy(old) Traceback (most recent call last): ... NotImplementedError: Deep copying of Nodes objects is not supported, as it would require to make deep copies of the Node objects themselves, which is in conflict with using their names as identifiers.
- class hydpy.core.devicetools.Nodes(*values: MayNonerable2[Node, str], mutable: bool = True, defaultvariable: NodeVariableType = 'Q')[source]¶
-
A container class for handling
Nodeobjects.For the general usage of
Nodesobjects, please see the documentation on its base classDevices.Class
Nodesprovides the additional keyword argument defaultvariable. Use it to temporarily change the default variable “Q” to another value during the initialisation of newNodeobjects:>>> from hydpy import Nodes >>> a1, t2 = Nodes("a1", "a2", defaultvariable="A") >>> a1 Node("a1", variable="A")
Be aware that changing the default variable does not affect already existing nodes:
>>> a1, b1 = Nodes("a1", "b1", defaultvariable="B") >>> a1 Node("a1", variable="A") >>> b1 Node("b1", variable="B")
- prepare_allseries(allocate_ram: bool = True, jit: bool = False) None[source]¶
Call method
prepare_allseries()of all handledNodeobjects.
- prepare_simseries(allocate_ram: bool = True, read_jit: bool = False, write_jit: bool = False) None[source]¶
Call method
prepare_simseries()of all handledNodeobjects.
- prepare_obsseries(allocate_ram: bool = True, read_jit: bool = False, write_jit: bool = False) None[source]¶
Call method
prepare_obsseries()of all handledNodeobjects.
- load_allseries() None[source]¶
Call methods
load_simseries()andload_obsseries().
- load_simseries() None[source]¶
Call method
load_series()of allSimobjects with an activatedmemoryflag.
- load_obsseries() None[source]¶
Call method
load_series()of allObsobjects with an activatedmemoryflag.
- save_allseries() None[source]¶
Call methods
save_simseries()andsave_obsseries().
- save_simseries() None[source]¶
Call method
save_series()of allSimobjects with an activatedmemoryflag.
- save_obsseries() None[source]¶
Call method
save_series()of allObsobjects with an activatedmemoryflag.
- class hydpy.core.devicetools.Elements(*values: TypeDevice | str | Iterable[TypeDevice | str] | None, mutable: bool = True)[source]¶
-
A container for handling
Elementobjects.For the general usage of
Elementsobjects, please see the documentation on its base classDevices.- property collectives: dict[str | None, tuple[Element, ...]]¶
The names and members of all currently relevant collectives.
Note that all
Elementinstances not belonging to anycollectiveare returned as a separate group:>>> from hydpy import Element, Elements >>> Elements().collectives {} >>> for group, elements in Elements( ... Element("a"), Element("b1", collective="b"), Element("c"), ... Element("d1", collective="d"), Element("b2", collective="b") ... ).collectives.items(): ... print(group, [e.name for e in elements]) None ['a', 'c'] b ['b1', 'b2'] d ['d1']
- unite_collectives() Elements[source]¶
Create overarching elements for all original elements that belong to a collective.
All elements of the same
collectivemust be handled as one entity during simulation. A typical use case is that individual elements describe different channels of a large river network, and all of them must be handled simultaneously by a single routing model instance to account for backwater effects. We create such an example by combining instances ofmusk_classic(for “hydrological” routing neglecting backwater effects) andsw1d_channel(for “hydrodynamic” routing considering backwater effects).First, we create a
FusedVariableobject for connecting the inlets and outlets ofmusk_classicandsw1d_channel:>>> from hydpy import FusedVariable >>> from hydpy.aliases import (musk_inlets_Q, sw1d_inlets_LongQ, ... musk_outlets_Q, sw1d_outlets_LongQ) >>> q = FusedVariable("Q", musk_inlets_Q, sw1d_inlets_LongQ, ... musk_outlets_Q, sw1d_outlets_LongQ)
The spatial setting is more concise than realistic and consists of four channels. Channel A discharges into channel B, which discharges into channel C, which discharges into channel D. We neglect backwater effects within channels A and D. Hence we do not need to associate them with a collective and
musk_classicbecomes an appropriate choice. Channel B and C are represented by separate collectives. Hence, the setting could account for backwater effects within both channels but not between them. Channel B consists only of a single subchannel (represented by element b), while channel C consists of two subchannels (represented by elements c1 and c2):>>> from hydpy import Element, Elements, Nodes >>> q_a, q_a_b, q_b_c1, q_c1_c2, q_c2_d, q_d = Nodes( ... "q_a", "q_a_b", "q_b_c1", "q_c1_c2", "q_c2_d", "q_d", ... defaultvariable=q) >>> e_a = Element("e_a", inlets=q_a, outlets=q_a_b) >>> e_b = Element("e_b", collective="B", inlets=q_a_b, outlets=q_b_c1) >>> e_c1 = Element("e_c1", collective="C", inlets=q_b_c1, outlets=q_c1_c2) >>> e_c2 = Element("e_c2", collective="C", inlets=q_c1_c2, outlets=q_c2_d) >>> e_d = Element("e_d", inlets=q_c2_d, outlets=q_d) >>> elements = Elements(e_a, e_b, e_c1, e_c2, e_d)
Method
unite_collectives()expects only those elements belonging to a collective to come with a readyModelinstance. So we only need to preparesw1d_channelinstances for elements b, c1, and c2, including the required submodels:>>> from hydpy import prepare_model, pub >>> pub.timegrids = "2000-01-01", "2000-01-02", "1d" >>> for element in (e_b, e_c1, e_c2): ... channel = prepare_model("sw1d_channel") ... channel.parameters.control.nmbsegments(1) ... add_storage = channel.add_storagemodel_v1 ... with add_storage("sw1d_storage", position=0, update=False): ... pass ... if element in (e_b, e_c1): ... with channel.add_routingmodel_v1("sw1d_q_in", position=0): ... pass ... if element is e_c1: ... with channel.add_routingmodel_v2("sw1d_lias", position=1): ... lengthupstream(1.0) ... lengthdownstream(1.0) ... if element in (e_b, e_c2): ... with channel.add_routingmodel_v3("sw1d_weir_out", position=1): ... pass ... element.model = channel
Based on the defined five elements, method
unite_collectives()returns four:>>> elements.unite_collectives() Elements("B", "C", "e_a", "e_d")
The returned elements a and d are the same as those defined initially, as they do not belong to any collectives:
>>> collectives = elements.unite_collectives() >>> collectives.e_a is e_a True
However, the elements B and C are new. B replaces element b, and C replaces elements c1 and c2. Both handle instances of
sw1d_network, which is the suitable model for connecting and applying the submodels ofsw1d_channel(seeModelCoupler):>>> e_b, e_c = collectives.B, collectives.C >>> e_b.model.name 'sw1d_network'
The new element B has the same inlet and outlet nodes as b:
>>> e_b Element("B", inlets="q_a_b", outlets="q_b_c1")
However, C adopts both outlet nodes of c1 and c2 but only the inlet node of c1, which is relevant for clarifying the
deviceorderduring simulations:>>> e_c Element("C", inlets="q_b_c1", outlets=["q_c1_c2", "q_c2_d"])
The following technical checks ensure the underlying coupling mechanisms actually worked:
>>> assert e_b.model.storagemodels.number == 1 >>> assert e_c.model.storagemodels.number == 2 >>> assert e_b.model.routingmodels.number == 2 >>> assert e_c.model.routingmodels.number == 3 >>> assert e_c.model.routingmodels[1].routingmodelsdownstream[0] is e_c.model.routingmodels[2]
unite_collectives()raises the following error if an element belonging to a collective does not handle aModelinstance:>>> e_d.collective = "D" >>> elements.unite_collectives() Traceback (most recent call last): ... hydpy.core.exceptiontools.AttributeNotReady: While trying to unite the elements belonging to collective `D`, the following error occurred: The model object of element `e_d` has been requested but not been prepared so far.
unite_collectives()raises the following error if an element belonging to a collective does handle an unsuitableModelinstance:>>> e_d.model = prepare_model("musk_classic") >>> elements.unite_collectives() Traceback (most recent call last): ... TypeError: While trying to unite the elements belonging to collective `D`, the following error occurred: Model `musk_classic` of element `e_d` does not provide a function for coupling models that belong to the same collective.
- prepare_models() None[source]¶
Call method
prepare_model()of all handleElementobjects.We show, based on the HydPy-H-Lahn example project, that method
init_model()prepares theModelobjects of all elements, including building the required connections and updating the derived parameters:>>> from hydpy.core.testtools import prepare_full_example_1 >>> prepare_full_example_1() >>> from hydpy import attrready, HydPy, pub, TestIO >>> with TestIO(): ... hp = HydPy("HydPy-H-Lahn") ... pub.timegrids = "1996-01-01", "1996-02-01", "1d" ... hp.prepare_network() ... hp.prepare_models() >>> hp.elements.land_dill_assl.model.parameters.derived.dt dt(0.000833)
Wrong control files result in error messages like the following:
>>> with TestIO(): ... with open("HydPy-H-Lahn/control/default/land_dill_assl.py", ... "a") as file_: ... _ = file_.write("zonetype(-1)") ... hp.prepare_models() Traceback (most recent call last): ... ValueError: While trying to initialise the model object of element `land_dill_assl`, the following error occurred: While trying to load the control file `...land_dill_assl.py`, the following error occurred: At least one value of parameter `zonetype` of element `?` is not valid.
By default, missing control files result in exceptions:
>>> del hp.elements.land_dill_assl.model >>> import os >>> with TestIO(): ... os.remove("HydPy-H-Lahn/control/default/land_dill_assl.py") ... hp.prepare_models() Traceback (most recent call last): ... FileNotFoundError: While trying to initialise the model object of element `land_dill_assl`, the following error occurred: While trying to load the control file `...land_dill_assl.py`, the following error occurred: ... >>> attrready(hp.elements.land_dill_assl, "model") False
When building new, still incomplete HydPy projects, this behaviour can be annoying. After setting the option
warnmissingcontrolfiletoFalse, missing control files result in a warning only:>>> with TestIO(): ... with pub.options.warnmissingcontrolfile(True): ... hp.prepare_models() Traceback (most recent call last): ... UserWarning: Due to a missing or no accessible control file, no model could be initialised for element `land_dill_assl` >>> attrready(hp.elements.land_dill_assl, "model") False
- init_models() None[source]¶
Deprecated: use method
prepare_models()instead.>>> from hydpy import Elements >>> from unittest import mock >>> with mock.patch.object(Elements, "prepare_models") as mocked: ... elements = Elements() ... elements.init_models() Traceback (most recent call last): ... hydpy.core.exceptiontools.HydPyDeprecationWarning: Method `init_models` of class `Elements` is deprecated. Use method `prepare_models` instead. >>> mocked.call_args_list [call()]
- save_controls(parameterstep: timetools.PeriodConstrArg | None = None, simulationstep: timetools.PeriodConstrArg | None = None, auxfiler: auxfiletools.Auxfiler | None = None) None[source]¶
Save the control parameters of the
Modelobject handled by eachElementobject and eventually the ones handled by the givenAuxfilerobject.
- update_parameters() None[source]¶
Update the derived parameters of all models managed by the respective elements.
- load_conditions() None[source]¶
Load the initial conditions of the
Modelobject handled by eachElementobject.
- save_conditions() None[source]¶
Save the calculated conditions of the
Modelobject handled by eachElementobject.
- trim_conditions() None[source]¶
Call method
trim_conditions()of theModelobject handled by eachElementobject.
- reset_conditions() None[source]¶
Call method
reset_conditions()of theModelobject handled by eachElementobject.
- property conditions: dict[str, dict[str, dict[str, dict[str, float | ndarray[tuple[Any, ...], dtype[float64]]]]]]¶
A nested dictionary that contains the values of all
ConditionSequenceobjects of all currently handled models.See the documentation on property
conditionsfor further information.
- prepare_allseries(allocate_ram: bool = True, jit: bool = False) None[source]¶
Call method
prepare_allseries()of all handledElementobjects.
- prepare_inputseries(allocate_ram: bool = True, read_jit: bool = False, write_jit: bool = False) None[source]¶
Call method
prepare_inputseries()of all handledElementobjects.
- prepare_factorseries(allocate_ram: bool = True, write_jit: bool = False) None[source]¶
Call method
prepare_factorseries()of all handledElementobjects.
- prepare_fluxseries(allocate_ram: bool = True, write_jit: bool = False) None[source]¶
Call method
prepare_fluxseries()of all handledElementobjects.
- prepare_stateseries(allocate_ram: bool = True, write_jit: bool = False) None[source]¶
Call method
prepare_stateseries()of all handledElementobjects.
- prepare_linkseries(allocate_ram: bool = True, write_jit: bool = False) None[source]¶
Call method
prepare_linkseries()of all handledElementobjects.
- load_allseries() None[source]¶
Call method
load_inputseries()of all handledElementobjects.
- load_inputseries() None[source]¶
Call method
load_inputseries()of all handledElementobjects.
- load_factorseries() None[source]¶
Call method
load_factorseries()of all handledElementobjects.
- load_fluxseries() None[source]¶
Call method
load_fluxseries()of all handledElementobjects.
- load_stateseries() None[source]¶
Call method
load_stateseries()of all handledElementobjects.
- load_linkseries() None[source]¶
Call method
load_linkseries()of all handledElementobjects.
- save_allseries() None[source]¶
Call method
save_allseries()of all handledElementobjects.
- save_inputseries() None[source]¶
Call method
save_inputseries()of all handledElementobjects.
- save_factorseries() None[source]¶
Call method
save_factorseries()of all handledElementobjects.
- save_fluxseries() None[source]¶
Call method
save_fluxseries()of all handledElementobjects.
- save_stateseries() None[source]¶
Call method
save_stateseries()of all handledElementobjects.
- save_linkseries() None[source]¶
Call method
save_linkseries()of all handledElementobjects.
- class hydpy.core.devicetools.Device(value: Device | str, *args: object, **kwargs: object)[source]¶
Bases:
objectBase class for class
Elementand classNode.- classmethod query_all() Devices[Self][source]¶
Get all
NodeorElementobjects initialised so far.See the main documentation on module
devicetoolsfor further information.
- classmethod extract_new() Devices[Self][source]¶
Gather all “new”
NodeorElementobjects.See the main documentation on module
devicetoolsfor further information.
- classmethod clear_all() None[source]¶
Clear the registry from all initialised
NodeorElementobjects.See the main documentation on module
devicetoolsfor further information.
- property name: str¶
Name of the actual
NodeorElementobject.Device names serve as identifiers, as explained in the main documentation on module
devicetools. Hence, define them carefully:>>> from hydpy import Node >>> Node.clear_all() >>> node1, node2 = Node("n1"), Node("n2") >>> node1 is Node("n1") True >>> node1 is Node("n2") False
Each device name must be a valid variable identifier (see function
valid_variable_identifier()) to allow for attribute access:>>> from hydpy import Nodes >>> nodes = Nodes(node1, "n2") >>> nodes.n1 Node("n1", variable="Q")
Invalid variable identifiers result in errors like the following:
>>> node3 = Node("n 3") Traceback (most recent call last): ... ValueError: While trying to initialize a `Node` object with value `n 3` of type `str`, the following error occurred: The given name string `n 3` does not define a valid variable identifier. ...
When you change the name of a device (only do this for a good reason), the corresponding keys of all related
NodesandElementsobjects (as well as of the internal registry) change automatically:>>> Node.query_all() Nodes("n1", "n2") >>> node1.name = "n1a" >>> nodes Nodes("n1a", "n2") >>> Node.query_all() Nodes("n1a", "n2")
- keywords¶
Keywords describing the actual
NodeorElementobject.The keywords are contained within a
Keywordsobject:>>> from hydpy import Node >>> node = Node("n", keywords="word0") >>> node.keywords Keywords("word0")
Assigning new words does not overwrite already existing ones. You are allowed to add them individually or within iterable objects:
>>> node.keywords = "word1" >>> node.keywords = "word2", "word3" >>> node.keywords Keywords("word0", "word1", "word2", "word3")
Additionally, passing additional keywords to the constructor of class
NodeorElementworks also fine:>>> Node("n", keywords=("word3", "word4", "word5")) Node("n", variable="Q", keywords=["word0", "word1", "word2", "word3", "word4", "word5"])
You can delete all keywords at once:
>>> del node.keywords >>> node.keywords Keywords()
- class hydpy.core.devicetools.Node(value: Device | str, *args: object, **kwargs: object)[source]¶
Bases:
DeviceHandles the data flow between
Elementobjects.Nodeobjects always handle two sequences, aSimobject for simulated values and anObsobject for measured values:>>> from hydpy import Node >>> node = Node("test") >>> for sequence in node.sequences: ... print(sequence) sim(0.0) obs(0.0)
Each node can handle an arbitrary number of “input” and “output” elements, available as instance attributes entries and exits, respectively:
>>> node.entries Elements() >>> node.exits Elements()
You cannot (or at least should not) add new elements manually:
>>> node.entries = "element" Traceback (most recent call last): ... AttributeError: ... >>> node.exits.add_device("element") Traceback (most recent call last): ... RuntimeError: While trying to add the device `element` to a Elements object, the following error occurred: Adding devices to immutable Elements objects is not allowed.
Instead, see the documentation on class
Elementon how to connectNodeandElementobjects properly.- masks = defaultmask of module hydpy.core.masktools¶
- sequences: sequencetools.NodeSequences¶
- property entries: Elements¶
Group of
Elementobjects which set the the simulated value of theNodeobject.
- property exits: Elements¶
Group of
Elementobjects that query the simulated or observed value of the actualNodeobject.
- property variable: NodeVariableType¶
The variable handled by the actual
Nodeobject.By default, we suppose that nodes route discharge:
>>> from hydpy import Node >>> node = Node("test1") >>> node.variable 'Q'
Each other string, as well as each
InputSequencesubclass, is acceptable (for further information, see the documentation on methodconnect()):>>> Node("test2", variable="H") Node("test2", variable="H") >>> from hydpy.models.hland.hland_inputs import T >>> Node("test3", variable=T) Node("test3", variable=hland_inputs_T)
The last example above shows that the string representations of nodes handling “class variables” use the aliases importable from the top level of the HydPy package:
>>> from hydpy.aliases import hland_inputs_P >>> Node("test4", variable=hland_inputs_P) Node("test4", variable=hland_inputs_P)
For some complex HydPy projects, one may need to fall back on
FusedVariableobjects. The string representation then relies on the name of the fused variable:>>> from hydpy import FusedVariable >>> from hydpy.aliases import lland_inputs_Nied >>> Precipitation = FusedVariable("Precip", hland_inputs_P, lland_inputs_Nied) >>> Node("test5", variable=Precipitation) Node("test5", variable=Precip)
To avoid confusion, one cannot change
variable:>>> node.variable = "H" Traceback (most recent call last): ... AttributeError: ... >>> Node("test1", variable="H") Traceback (most recent call last): ... ValueError: The variable to be represented by a Node instance cannot be changed. The variable of node `test1` is `Q` instead of `H`. Keep in mind, that `name` is the unique identifier of node objects.
- property deploymode: Literal['newsim', 'oldsim', 'obs', 'obs_newsim', 'obs_oldsim', 'oldsim_bi', 'obs_bi', 'obs_oldsim_bi', 'newsim_update', 'obs_newsim_update']¶
Defines the kind of information a node offers its exit elements, eventually, its entry elements.
HydPy supports the following user-relevant modes:
newsim: Deploy the simulated values calculated just recently. newsim is the default mode, used, for example, when a node receives a discharge value from an upstream element and passes it to the downstream element directly.
obs: Deploy observed values instead of simulated values. The node still receives the simulated values from its upstream element(s). However, it deploys values to its downstream element(s), which are defined externally. Usually, these values are observations made available within a time series file. See the documentation on module
sequencetoolsfor further information on file specifications.oldsim: Similar to mode obs. However, it is usually applied when a node is supposed to deploy simulated values that have been calculated in a previous simulation run and stored in a sequence file.
obs_newsim: Combination of mode obs and newsim. Mode obs_newsim gives priority to the provision of observation values. New simulation values serve as a replacement for missing observed values.
obs_oldsim: Combination of mode obs and oldsim. Mode obs_oldsim gives priority to the provision of observation values. Old simulation values serve as a replacement for missing observed values.
obs_bi: Similar to the obs mode but triggers “bidirectional” deployment. All bidirectional modes only apply if the upstream element(s) do not calculate data for but expect from their downstream nodes. A typical example is using discharge measurements as lower boundary conditions for a hydrodynamical flood routing method.
oldsim_bi: The bidirectional version of the oldsim mode.
obs_oldsim_bi: The bidirectional version of the obs_oldsim mode.
One relevant difference between modes obs and oldsim is that the external values are either handled by the obs or the sim sequence object. Hence, if you select the oldsim mode, the values of the upstream elements calculated within the current simulation are not available (e.g. for parameter calibration) after the simulation finishes.
There are also the “update” modes newsim_update and obs_newsim_update. HydPy sets these modes temporarily when a newsim or a obs_newsim node represents a transition between a network’s parallelisable and non-parallelisable parts. Users should activate these two modes only for testing purposes.
Please refer to the documentation on method
simulate()of classHydPy, which provides some application examples.>>> from hydpy import Node >>> node = Node("test") >>> node.deploymode 'newsim' >>> node.deploymode = "obs" >>> node.deploymode 'obs' >>> node.deploymode = "oldsim" >>> node.deploymode 'oldsim' >>> node.deploymode = "obs_newsim" >>> node.deploymode 'obs_newsim' >>> node.deploymode = "obs_oldsim" >>> node.deploymode 'obs_oldsim' >>> node.deploymode = "oldsim_bi" >>> node.deploymode 'oldsim_bi' >>> node.deploymode = "obs_bi" >>> node.deploymode 'obs_bi' >>> node.deploymode = "obs_oldsim_bi" >>> node.deploymode 'obs_oldsim_bi' >>> node.deploymode = "newsim_update" >>> node.deploymode 'newsim_update' >>> node.deploymode = "obs_newsim_update" >>> node.deploymode 'obs_newsim_update' >>> node.deploymode = "newsim" >>> node.deploymode 'newsim' >>> node.deploymode = "oldobs" Traceback (most recent call last): ... ValueError: When trying to set the routing mode of node `test`, the value `oldobs` was given, but only the following values are allowed: `newsim`, `oldsim`, `obs`, `obs_newsim`, `obs_oldsim`, `oldsim_bi`, `obs_bi`, `obs_oldsim_bi`, `newsim_update`, and `obs_newsim_update`.
- get_double(group: Literal['inlets', 'outlets', 'observers', 'receivers', 'senders', 'inputs', 'outputs']) Double[source]¶
Return the
Doubleobject appropriate for the givenElementinput or output group and the actualdeploymode.Method
get_double()should interest framework developers only (and eventually model developers).Let
Nodeobject node1 handle different simulation and observation values:>>> from hydpy import Node >>> node = Node("node1") >>> node.sequences.sim = 1.0 >>> node.sequences.obs = 2.0
The following test function shows for a given
deploymodeif methodget_double()either returns theDoubleobject handling the simulated value (1.0) or the one handling the observed value (2.0):>>> def test(deploymode): ... node.deploymode = deploymode ... for group in ( "inlets", "observers", "receivers", "inputs"): ... end = None if group == "inputs" else ", " ... print(group, node.get_double(group), sep=": ", end=end) ... for group in ("outlets", "senders", "outputs"): ... end = None if group == "outputs" else ", " ... print(group, node.get_double(group), sep=": ", end=end)
In the default mode, nodes (passively) route simulated values by offering the
Doubleobject of sequenceSimto allElementinput and output groups:>>> test("newsim") inlets: 1.0, observers: 1.0, receivers: 1.0, inputs: 1.0 outlets: 1.0, senders: 1.0, outputs: 1.0
Setting
deploymodeto obs means that a node receives simulated values but provides observed values:>>> test("obs") inlets: 2.0, observers: 2.0, receivers: 2.0, inputs: 2.0 outlets: 1.0, senders: 1.0, outputs: 1.0
With
deploymodeset to oldsim, the node provides (previously) simulated values but does not receive any. Methodget_double()returns a dummyDoubleobject initialised to 0.0 in this case:>>> test("oldsim") inlets: 1.0, observers: 1.0, receivers: 1.0, inputs: 1.0 outlets: 0.0, senders: 0.0, outputs: 0.0
For obs_newsim, the result is like for obs because, for missing data, HydPy temporarily copies newly calculated values into the observation sequence during simulation:
>>> test("obs_newsim") inlets: 2.0, observers: 2.0, receivers: 2.0, inputs: 2.0 outlets: 1.0, senders: 1.0, outputs: 1.0
Similar holds for the obs_oldsim mode, but here
get_double()must ensure newly calculated values do not overwrite the “old” ones:>>> test("obs_oldsim") inlets: 2.0, observers: 2.0, receivers: 2.0, inputs: 2.0 outlets: 0.0, senders: 0.0, outputs: 0.0
All “bidirectional” modes require symmetrical connections, as they long for passing the same information in the downstream and the upstream direction:
>>> test("obs_bi") inlets: 2.0, observers: 2.0, receivers: 2.0, inputs: 2.0 outlets: 2.0, senders: 2.0, outputs: 2.0 >>> test("oldsim_bi") inlets: 1.0, observers: 1.0, receivers: 1.0, inputs: 1.0 outlets: 1.0, senders: 1.0, outputs: 1.0 >>> test("obs_oldsim_bi") inlets: 2.0, observers: 2.0, receivers: 2.0, inputs: 2.0 outlets: 2.0, senders: 2.0, outputs: 2.0
- reset(idx: int = 0) None[source]¶
Reset the actual value of the simulation sequence to zero.
>>> from hydpy import Node >>> node = Node("node1") >>> node.sequences.sim = 1.0 >>> node.reset() >>> node.sequences.sim sim(0.0)
- prepare_allseries(allocate_ram: bool = True, jit: bool = False) None[source]¶
Call method
prepare_simseries()with write_jit=jit and methodprepare_obsseries()with read_jit=jit.
- prepare_simseries(allocate_ram: bool = True, read_jit: bool = False, write_jit: bool = False) None[source]¶
Call method
prepare_series()of theSimsequence object.
- prepare_obsseries(allocate_ram: bool = True, read_jit: bool = False, write_jit: bool = False) None[source]¶
Call method
prepare_series()of theObssequence object.
- plot_allseries(*, labels: tuple[str, str] | None = None, colors: str | tuple[str, str] | None = None, linestyles: Literal['-', '--', '-.', ':', 'solid', 'dashed', 'dashdot', 'dotted'] | tuple[Literal['-', '--', '-.', ':', 'solid', 'dashed', 'dashdot', 'dotted'], Literal['-', '--', '-.', ':', 'solid', 'dashed', 'dashdot', 'dotted']] | None = None, linewidths: int | tuple[int, int] | None = None, focus: bool = False, stepsize: Literal['daily', 'd', 'monthly', 'm', 'yearly', 'y'] | None = None) Figure[source]¶
Plot the
seriesdata of both theSimand theObssequence object.We demonstrate the functionalities of method
plot_allseries()based on the Lahn example project:>>> from hydpy.core.testtools import prepare_full_example_2 >>> hp, pub, _ = prepare_full_example_2(lastdate="1997-01-01")
We perform a simulation run and calculate “observed” values for node dill_assl:
>>> hp.simulate() >>> dill_assl = hp.nodes.dill_assl >>> dill_assl.sequences.obs.series = dill_assl.sequences.sim.series + 10.0
A call to method
plot_allseries()prints the time series of both sequences to the screen immediately (if not, you need to activate the interactive mode of matplotlib first):>>> figure = dill_assl.plot_allseries()
Subsequent calls to
plot_allseries()or the related methodsplot_simseries()andplot_obsseries()of nodes add further time series data to the existing plot:>>> lahn_marb = hp.nodes.lahn_marb >>> figure = lahn_marb.plot_simseries()
You can modify the appearance of the lines by passing different arguments:
>>> lahn_marb.sequences.obs.series = lahn_marb.sequences.sim.series + 10.0 >>> figure = lahn_marb.plot_obsseries(color="black", linestyle="dashed")
All mentioned plotting functions return a
matplotlibFigureobject. Use it for further plot handling, e.g. adding a title and saving the current figure to disk:>>> from hydpy.core.testtools import save_autofig >>> text = figure.axes[0].set_title('daily') >>> save_autofig("Node_plot_allseries_1.png", figure)
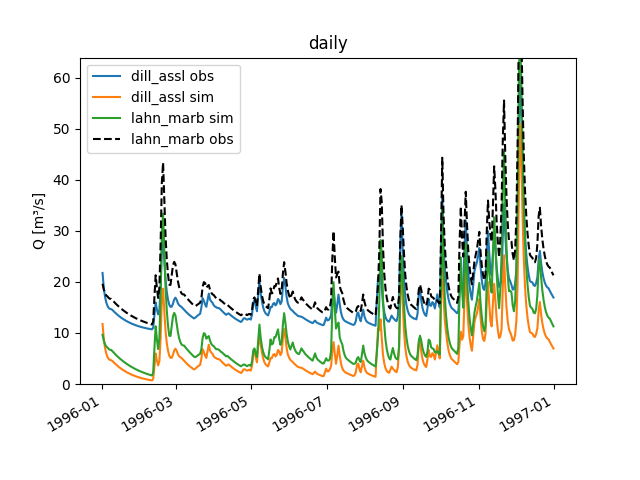
You can plot the data in an aggregated manner (see the documentation on the function
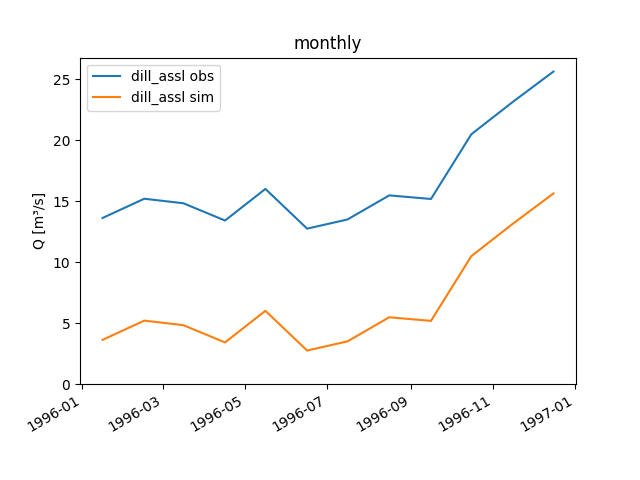
aggregate_series()for the supported step sizes and further details):>>> figure = dill_assl.plot_allseries(stepsize="monthly") >>> text = figure.axes[0].set_title('monthly') >>> save_autofig("Node_plot_allseries_2.png", figure)
You can restrict the plotted period via the
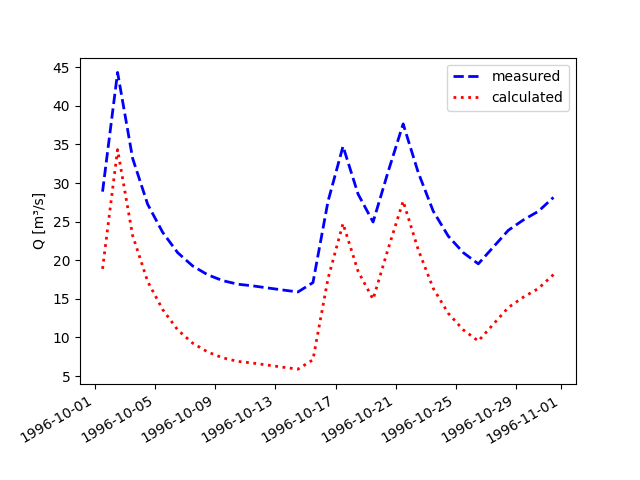
eval_Timegridand overwrite the time series label and other defaults via keyword arguments. For tuples passed to methodplot_allseries(), the first entry corresponds to the observation and the second one to the simulation results:>>> pub.timegrids.eval_.dates = "1996-10-01", "1996-11-01" >>> figure = lahn_marb.plot_allseries(labels=("measured", "calculated"), ... colors=("blue", "red"), ... linewidths=2, ... linestyles=("--", ":"), ... focus=True,) >>> save_autofig("Node_plot_allseries_3.png", figure)
When necessary, all plotting methods raise errors like the following:
>>> figure = lahn_marb.plot_allseries(stepsize="quaterly") Traceback (most recent call last): ... ValueError: While trying to plot the time series of the sequences obs and sim of node `lahn_marb` for the period `1996-10-01 00:00:00` to `1996-11-01 00:00:00`, the following error occurred: While trying to aggregate the given series, the following error occurred: Argument `stepsize` received value `quaterly`, but only the following ones are supported: `monthly` (default), `daily`, and `yearly`.
>>> from hydpy import pub >>> del pub.timegrids >>> figure = lahn_marb.plot_allseries() Traceback (most recent call last): ... hydpy.core.exceptiontools.AttributeNotReady: While trying to plot the time series of the sequences obs and sim of node `lahn_marb`, the following error occurred: Attribute timegrids of module `pub` is not defined at the moment.
- plot_simseries(*, label: str | None = None, color: str | None = None, linestyle: Literal['-', '--', '-.', ':', 'solid', 'dashed', 'dashdot', 'dotted'] | None = None, linewidth: int | None = None, focus: bool = False, stepsize: Literal['daily', 'd', 'monthly', 'm', 'yearly', 'y'] | None = None) Figure[source]¶
Plot the
seriesof theSimsequence object.See method
plot_allseries()for further information.
- plot_obsseries(*, label: str | None = None, color: str | None = None, linestyle: Literal['-', '--', '-.', ':', 'solid', 'dashed', 'dashdot', 'dotted'] | None = None, linewidth: int | None = None, focus: bool = False, stepsize: Literal['daily', 'd', 'monthly', 'm', 'yearly', 'y'] | None = None) Figure[source]¶
Plot the
seriesof theObssequence object.See method
plot_allseries()for further information.
- plot_alldurationcurves(*, labels: tuple[str, str] | None = None, colors: str | tuple[str, str] | None = None, linestyles: Literal['-', '--', '-.', ':', 'solid', 'dashed', 'dashdot', 'dotted'] | tuple[Literal['-', '--', '-.', ':', 'solid', 'dashed', 'dashdot', 'dotted'], Literal['-', '--', '-.', ':', 'solid', 'dashed', 'dashdot', 'dotted']] | None = None, linewidths: int | tuple[int, int] | None = None, logscale: bool = True) Figure[source]¶
Plot the
evalseriesof both theSimand theObssequence in the form of a (flow) duration curve.Method
plot_alldurationcurves()works very similar to methodplot_allseries(). Hence, we restrict the following examples to their key differences.We use the Lahn example project in the same way:
>>> from hydpy.core.testtools import prepare_full_example_2 >>> hp, pub, _ = prepare_full_example_2(lastdate="1997-01-01") >>> hp.simulate() >>> dill_assl = hp.nodes.dill_assl >>> dill_assl.sequences.obs.series = dill_assl.sequences.sim.series + 10.0
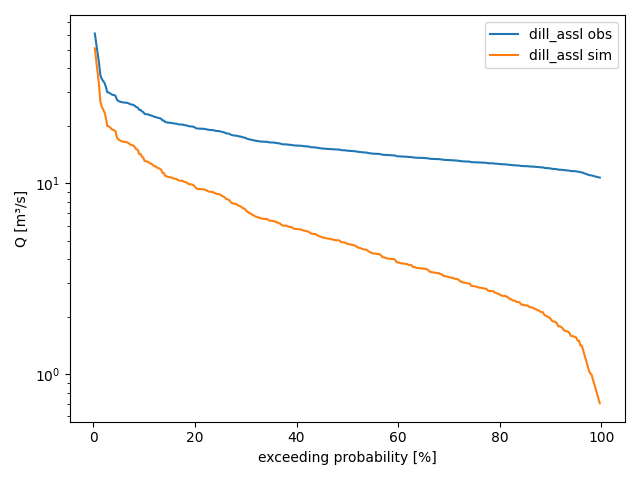
By default, method
plot_alldurationcurves()chooses a logarithmic y-scale and the following graphical settings:>>> figure = dill_assl.plot_alldurationcurves() >>> from hydpy.core.testtools import save_autofig >>> save_autofig("Node_plot_alldurationcurves_1.png", figure)
Despite its option to disable the logarithmic y-scale, the configuration works as for method
plot_allseries():>>> figure = dill_assl.plot_alldurationcurves( ... labels=("observed", "simulated"), ... colors=("black", "green"), ... linestyles=":", ... linewidths=(1, 2), ... logscale=False, ... ) >>> _ = figure.gca().set_title("Asslar (Dill)") >>> figure.tight_layout() >>> save_autofig("Node_plot_alldurationcurves_2.png", figure)
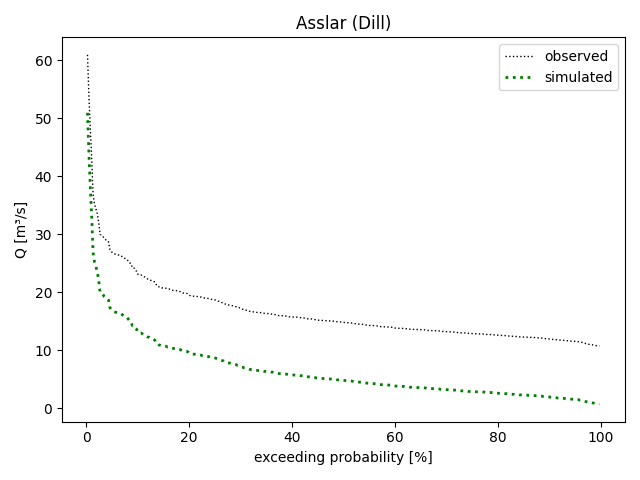
Again, you can modify the considered period via the
eval_time grid and plot the different curves individually via methodsplot_obsdurationcurve()andplot_simdurationcurve()(in which case the last method call decides whether the y-scale is logarithmic or linear):>>> pub.timegrids.eval_.lastdate = "1996-06-01" >>> figure = dill_assl.plot_obsdurationcurve() >>> figure = dill_assl.plot_simdurationcurve( ... label="observed", ... color="green", ... linestyle=":", ... linewidth=2, ... logscale=False, ... ) >>> save_autofig("Node_plot_alldurationcurves_3.png", figure)
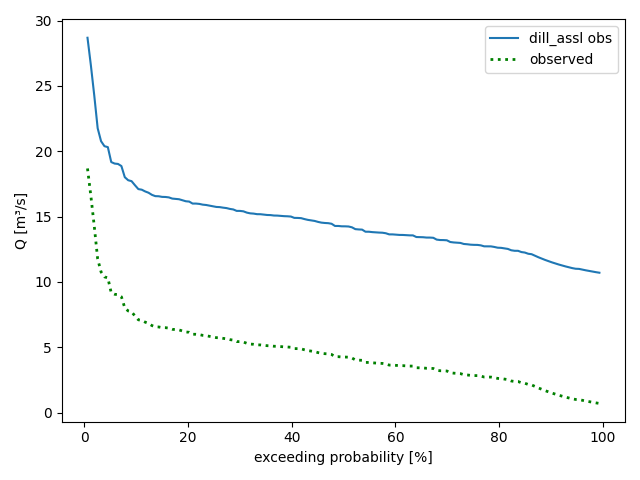
Misspecifications result in error messages like the following:
>>> dill_assl.plot_alldurationcurves(linestyles=".") Traceback (most recent call last): ... ValueError: While trying to plot the duration curves of the sequences obs and sim of node `dill_assl` for the period `1996-01-01 00:00:00` to `1996-06-01 00:00:00`, the following error occurred: '.' is not a valid value for ls; supported values are '-', '--', '-.', ':', 'None', ' ', '', 'solid', 'dashed', 'dashdot', 'dotted'
- plot_simdurationcurve(*, label: str | None = None, color: str | None = None, linestyle: Literal['-', '--', '-.', ':', 'solid', 'dashed', 'dashdot', 'dotted'] | None = None, linewidth: int | None = None, logscale: bool = True) Figure[source]¶
Plot the
seriesof theSimsequence object.See method
plot_allseries()for further information.
- plot_obsdurationcurve(*, label: str | None = None, color: str | None = None, linestyle: Literal['-', '--', '-.', ':', 'solid', 'dashed', 'dashdot', 'dotted'] | None = None, linewidth: int | None = None, logscale: bool = True) Figure[source]¶
Plot the duration curve of the
seriesof theObssequence object.See method
plot_allseries()for further information.
- class hydpy.core.devicetools.Element(value: Device | str, *args: object, **kwargs: object)[source]¶
Bases:
Device- Handles a
Modelobject and connects it to other models via Nodeobjects.When preparing
Elementobjects, one links them to nodes of different “groups”, each group of nodes implemented as an immutableNodesobject:inletsandoutletsnodes handle, for example, the inflow to and the outflow from the respective element.receivers,observers, andsendersnodes are thought for information flow.receiversnodes allow for arbitrarily directed connections, even to downstream locations (for example, to inform adammodel about the discharge at a gauge downstream). In contrast,observersnodes do not support such cyclic connections. However,observersnodes offer the advantage of direct information exchange, whilereceiversnodes provide their information with a delay of one simulation time step (to break possible cycles in the spatial simulation order).inputsnodes provide optional input information, for example, interpolated precipitation that could alternatively be read from files as well.outputsnodes query optional output information, for example, the water level of a dam.
You can select the relevant nodes either by passing them explicitly or passing their name both as single objects or as objects contained within an iterable object:
>>> from hydpy import Element, Node >>> Element("test", ... inlets="inl1", ... outlets=Node("outl1"), ... receivers=("rec1", Node("rec2"))) Element("test", inlets="inl1", outlets="outl1", receivers=["rec1", "rec2"])
Repeating such a statement with different nodes adds them to the existing ones without any conflict in case of repeated specifications:
>>> Element("test", ... inlets="inl1", ... observers="obs1", ... receivers=("rec2", "rec3"), ... senders="sen1", ... inputs="inp1", ... outputs="outp1") Element("test", inlets="inl1", outlets="outl1", observers="obs1", receivers=["rec1", "rec2", "rec3"], senders="sen1", inputs="inp1", outputs="outp1")
Subsequent adding of nodes also works via property access:
>>> test = Element("test") >>> test.inlets = "inl2" >>> test.outlets = None >>> test.observers = "obs1" >>> test.receivers = () >>> test.senders = "sen2", Node("sen3") >>> test.inputs = [] >>> test.outputs = Node("outp2") >>> test Element("test", inlets=["inl1", "inl2"], outlets="outl1", observers="obs1", receivers=["rec1", "rec2", "rec3"], senders=["sen1", "sen2", "sen3"], inputs="inp1", outputs=["outp1", "outp2"])
The properties try to verify that all connections make sense. For example, an element should never handle an inlet node that is also handles as an outlet, input, or output node:
>>> def try_impossible_combinations(group, *sequences): ... for sequence in sequences: ... try: ... setattr(test, group, sequence) ... except ValueError as error: ... print(error) ... else: ... assert False
>>> try_impossible_combinations("inlets", "outl1", "sen1", "outp1") For element `test`, the given inlet node `outl1` is already defined as an outlet node, which is not allowed. For element `test`, the given inlet node `sen1` is already defined as a sender node, which is not allowed. For element `test`, the given inlet node `outp1` is already defined as an output node, which is not allowed.
Similar holds for the outlet nodes:
>>> try_impossible_combinations("outlets", "inl1", "obs1", "inp1", "outp1") For element `test`, the given outlet node `inl1` is already defined as an inlet node, which is not allowed. For element `test`, the given outlet node `obs1` is already defined as an observer node, which is not allowed. For element `test`, the given outlet node `inp1` is already defined as an input node, which is not allowed. For element `test`, the given outlet node `outp1` is already defined as an output node, which is not allowed.
The following restrictions hold for the sender nodes:
>>> try_impossible_combinations("senders", "inl1", "obs1", "inp1", "outp1") For element `test`, the given sender node `inl1` is already defined as an inlet node, which is not allowed. For element `test`, the given sender node `obs1` is already defined as an observer node, which is not allowed. For element `test`, the given sender node `inp1` is already defined as an input node, which is not allowed. For element `test`, the given sender node `outp1` is already defined as an output node, which is not allowed.
The following restrictions hold for the observer nodes:
>>> try_impossible_combinations("observers", "outl1", "sen1", "outp1") For element `test`, the given observer node `outl1` is already defined as an outlet node, which is not allowed. For element `test`, the given observer node `sen1` is already defined as a sender node, which is not allowed. For element `test`, the given observer node `outp1` is already defined as an output node, which is not allowed.
The following restrictions hold for the input nodes:
>>> try_impossible_combinations("inputs", "outl1", "sen1", "outp1") For element `test`, the given input node `outl1` is already defined as an outlet node, which is not allowed. For element `test`, the given input node `sen1` is already defined as a sender node, which is not allowed. For element `test`, the given input node `outp1` is already defined as an output node, which is not allowed.
The following restrictions hold for the output nodes:
>>> try_impossible_combinations("outputs", "inl1", "obs1", "inp1", "outl1", "sen1") For element `test`, the given output node `inl1` is already defined as an inlet node, which is not allowed. For element `test`, the given output node `obs1` is already defined as an observer node, which is not allowed. For element `test`, the given output node `inp1` is already defined as an input node, which is not allowed. For element `test`, the given output node `outl1` is already defined as an outlet node, which is not allowed. For element `test`, the given output node `sen1` is already defined as a sender node, which is not allowed.
Note that the discussed
Nodesobjects are immutable by default, disallowing to change them in other ways as described above:>>> test.inlets += "inl3" Traceback (most recent call last): ... RuntimeError: While trying to add the device `inl3` to a Nodes object, the following error occurred: Adding devices to immutable Nodes objects is not allowed.
Use the parameter force to change this behaviour:
>>> test.inlets.add_device("inl3", force=True)
However, it is up to you to make sure that the added node also handles the relevant element in the suitable group. In the discussed example, only node inl2 has been added properly but not node inl3:
>>> test.inlets.inl2.exits Elements("test") >>> test.inlets.inl3.exits Elements()
Some elements might belong to a
collective, which is a group of elements requiring simultaneous handling during simulation (see methodunite_collectives()). If needed, specify the collective’s name by the corresponding argument:>>> Element("part_1", collective="NileRiver", inlets="inl1") Element("part_1", collective="NileRiver", inlets="inl1")
The information persists when querying the same element from the internal registry, whether one specifies the collective’s name again or not:
>>> Element("part_1", collective="NileRiver") Element("part_1", collective="NileRiver", inlets="inl1")
>>> Element("part_1") Element("part_1", collective="NileRiver", inlets="inl1")
However, changing the collective via the constructor is forbidden as it might result in hard-to-find configuration errors:
>>> Element("part_1", collective="AmazonRiver") Traceback (most recent call last): ... RuntimeError: The collective name `AmazonRiver` is given, but element `part_1` is already a collective `NileRiver` member.
- inlets¶
Group of
Nodeobjects from which the handledModelobject queries its “upstream” input values (e.g. inflow).
- outlets¶
Group of
Nodeobjects to which the handledModelobject passes its “downstream” output values (e.g. outflow).
- observers¶
Group of non-downstream
Nodeobjects from which the handledModelobject queries its “remote” information values (e.g. inflow of a side-tributary into a downstream river).
- receivers¶
Group of arbitrarily placed
Nodeobjects from which the handledModelobject queries its “remote” information values (e.g. discharge at a downstream gauge).
- senders¶
Group of
Nodeobjects to which the handledModelobject passes its “remote” information values (e.g. water level of adammodel).
- inputs¶
Group of
Nodeobjects from which the handledModelobject queries its “external” input values instead of reading them from files (e.g. interpolated precipitation).
- outputs¶
Group of
Nodeobjects to which the handledModelobject passes its “internal” output values, available via sequences of typeFluxSequenceorStateSequence(e.g. potential evaporation).
- property model: modeltools.Model¶
The
Modelobject handled by the actualElementobject.Directly after their initialisation, elements do not know which model they require:
>>> from hydpy import attrready, Element >>> hland = Element("hland", outlets="outlet") >>> hland.model Traceback (most recent call last): ... hydpy.core.exceptiontools.AttributeNotReady: The model object of element `hland` has been requested but not been prepared so far.
During scripting and when working interactively in the Python shell, it is often convenient to assign a
Modeldirectly.>>> from hydpy.models.hland_96 import * >>> parameterstep("1d") >>> hland.model = model >>> hland.model.name 'hland_96'
>>> del hland.model >>> attrready(hland, "model") False
For the “usual” approach to preparing models, please see the method
prepare_model().The following examples show that assigning
Modelobjects to propertymodelcreates some connection required by the respective model type automatically. These examples should be relevant for developers only.The following
exch_branch_hbv96model branches a single input value (from to node inp) to multiple outputs (nodes out1 and out2):>>> from hydpy import Element, Node, reverse_model_wildcard_import, pub >>> reverse_model_wildcard_import() >>> pub.timegrids = "2000-01-01", "2000-01-02", "1d" >>> element = Element("a_branch", ... inlets="branch_input", ... outlets=("branch_output_1", "branch_output_2")) >>> inp = element.inlets.branch_input >>> out1, out2 = element.outlets >>> from hydpy.models.exch_branch_hbv96 import * >>> parameterstep() >>> delta(0.0) >>> minimum(0.0) >>> xpoints(0.0, 3.0) >>> ypoints(branch_output_1=[0.0, 1.0], branch_output_2=[0.0, 2.0]) >>> parameters.update() >>> element.model = model
To show that the inlet and outlet connections are built properly, we assign a new value to the inlet node inp and verify that the suitable fractions of this value are passed to the outlet nodes out1` and out2 by calling the method
simulate():>>> inp.sequences.sim = 999.0 >>> model.simulate(0) >>> fluxes.originalinput originalinput(999.0) >>> out1.sequences.sim sim(333.0) >>> out2.sequences.sim sim(666.0)
- prepare_model(clear_registry: bool = True) None[source]¶
Load the control file of the actual
Elementobject, initialise itsModelobject, build the required connections via (an eventually overridden version of) methodconnect()of classModel, and update its derived parameter values via calling (an eventually overridden version) of methodupdate()of classParameters.See method
prepare_models()of classHydPyand propertyModelof classElementfur further information.
- init_model(clear_registry: bool = True) None[source]¶
Deprecated: use method
prepare_model()instead.>>> from hydpy import Element >>> from unittest import mock >>> with mock.patch.object(Element, "prepare_model") as mocked: ... element = Element("test") ... element.init_model(False) Traceback (most recent call last): ... hydpy.core.exceptiontools.HydPyDeprecationWarning: Method `init_model` of class `Element` is deprecated. Use method `prepare_model` instead. >>> mocked.call_args_list [call(False)]
- property variables: set[NodeVariableType]¶
A set of all different
variablevalues of theNodeobjects directly connected to the actualElementobject.Suppose an element is connected to five nodes, which (partly) represent different variables:
>>> from hydpy import Element, Node >>> element = Element("Test", ... inlets=(Node("N1", "X"), Node("N2", "Y1")), ... outlets=(Node("N3", "X"), Node("N4", "Y2")), ... receivers=(Node("N5", "X"), Node("N6", "Y3")), ... senders=(Node("N7", "X"), Node("N8", "Y4")))
Property
variablesputs all the different variables of these nodes together:>>> sorted(element.variables) ['X', 'Y1', 'Y2', 'Y3', 'Y4']
- prepare_allseries(allocate_ram: bool = True, jit: bool = False) None[source]¶
Call method
prepare_allseries()of the currently handledModelinstance and its submodels.
- prepare_inputseries(allocate_ram: bool = True, read_jit: bool = False, write_jit: bool = False) None[source]¶
Call method
prepare_inputseries()of the currently handledModelinstance and its submodels.
- prepare_factorseries(allocate_ram: bool = True, write_jit: bool = False) None[source]¶
Call method
prepare_factorseries()of the currently handledModelinstance and its submodels.
- prepare_fluxseries(allocate_ram: bool = True, write_jit: bool = False) None[source]¶
Call method
prepare_fluxseries()of the currently handledModelinstance and its submodels.
- prepare_stateseries(allocate_ram: bool = True, write_jit: bool = False) None[source]¶
Call method
prepare_stateseries()of the currently handledModelinstance and its submodels.
- prepare_linkseries(allocate_ram: bool = True, write_jit: bool = False) None[source]¶
Call method
prepare_linkseries()of the currently handledModelinstance and its submodels.
- load_allseries() None[source]¶
Call method
load_allseries()of the currently handledModelinstance and its submodels.
- load_inputseries() None[source]¶
Call method
load_inputseries()of the currently handledModelinstance and its submodels.
- load_factorseries() None[source]¶
Call method
load_factorseries()of the currently handledModelinstance and its submodels.
- load_fluxseries() None[source]¶
Call method
load_fluxseries()of the currently handledModelinstance and its submodels.
- load_stateseries() None[source]¶
Call method
load_stateseries()of the currently handledModelinstance and its submodels.
- load_linkseries() None[source]¶
Call method
load_linkseries()of the currently handledModelinstance and its submodels.
- save_allseries() None[source]¶
Call method
save_allseries()of the currently handledModelinstance and its submodels.
- save_inputseries() None[source]¶
Call method
save_inputseries()of the currently handledModelinstance and its submodels.
- save_factorseries() None[source]¶
Call method
save_factorseries()of the currently handledModelinstance and its submodels.
- save_fluxseries() None[source]¶
Call method
save_fluxseries()of the currently handledModelinstance and its submodels.
- save_stateseries() None[source]¶
Call method
save_stateseries()of the currently handledModelinstance and its submodels.
- save_linkseries() None[source]¶
Call method
save_linkseries()of the currently handledModelinstance and its submodels.
- plot_inputseries(*sequences: IOSequence | type[IOSequence] | str, average: bool = False, labels: tuple[str, ...] | None = None, colors: str | tuple[str, ...] | None = None, linestyles: Literal['-', '--', '-.', ':', 'solid', 'dashed', 'dashdot', 'dotted'] | tuple[Literal['-', '--', '-.', ':', 'solid', 'dashed', 'dashdot', 'dotted'], ...] | None = None, linewidths: int | tuple[int, ...] | None = None, focus: bool = True) Figure[source]¶
Plot (the selected)
InputSequenceseriesvalues.We demonstrate the functionalities of method
plot_inputseries()based on the Lahn example project:>>> from hydpy.core.testtools import prepare_full_example_2 >>> hp, pub, _ = prepare_full_example_2(lastdate="1997-01-01")
Without any arguments,
plot_inputseries()prints the time series of all input sequences handled by its (sub)models directly to the screen (in our example,PandTofhland_96andNormalAirTemperatureandNormalEvapotranspirationofevap_pet_hbv96):>>> land = hp.elements.land_dill_assl >>> figure = land.plot_inputseries()
You can use the pyplot API of matplotlib to modify the returned figure or to save it to disk (or print it to the screen, in case the interactive mode of matplotlib is disabled):
>>> from hydpy.core.testtools import save_autofig >>> save_autofig("Element_plot_inputseries_complete.png", figure)
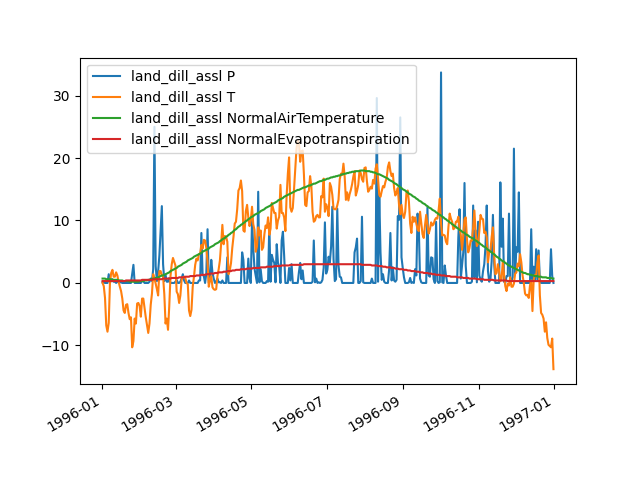
Select specific sequences by passing their names, types, or example objects:
>>> from hydpy.models.hland.hland_inputs import T >>> net = land.model.aetmodel.petmodel.sequences.inputs.normalevapotranspiration >>> figure = land.plot_inputseries("p", T, net) >>> save_autofig("Element_plot_inputseries_selection.png", figure)
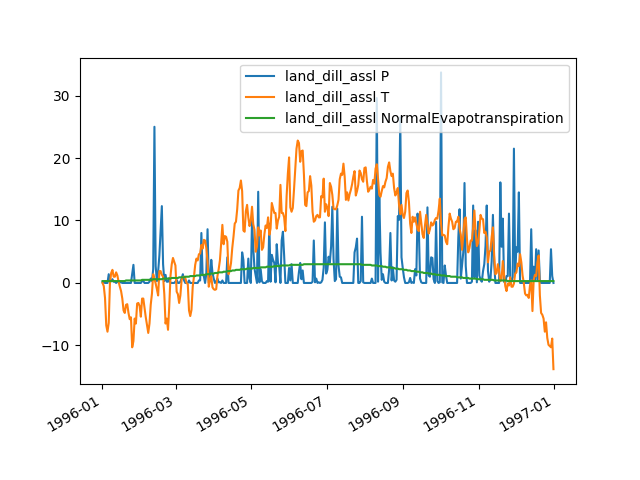
Misleading sequence specifiers result in the following error:
>>> figure = land.plot_inputseries("xy") Traceback (most recent call last): ... ValueError: No (sub)model handled by element `land_dill_assl` has an input sequence named `xy`.
Methods
plot_factorseries(),plot_fluxseries(), andplot_stateseries()work in the same manner. Before applying them, one has to calculate the time series of theFactorSequence,FluxSequence, andStateSequenceobjects:>>> hp.simulate()
The arguments “labels,” “colours,” “line styles,” and “line widths” can accept general or individual values:
>>> figure = land.plot_fluxseries( ... "q0", "q1", labels=("direct runoff", "base flow"), ... colors=("red", "green"), linestyles="--", linewidths=2) >>> save_autofig("Element_plot_fluxseries.png", figure)
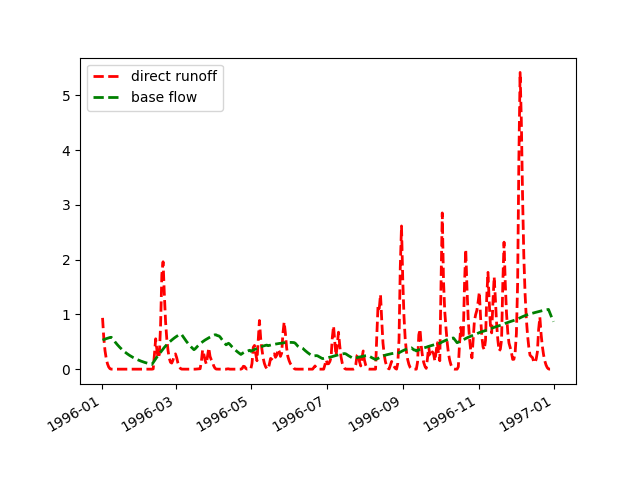
For 1- and 2-dimensional
IOSequenceobjects, all three methods plot the individual time series in the same colour. We demonstrate this for the frozen (SP) and the liquid (WC) water equivalent of the snow cover of different hydrological response units. Therefore, we restrict the shown period to February and March via theeval_time grid:>>> with pub.timegrids.eval_(firstdate="1996-02-01", lastdate="1996-04-01"): ... figure = land.plot_stateseries("sp", "wc") >>> save_autofig("Element_plot_stateseries.png", figure)
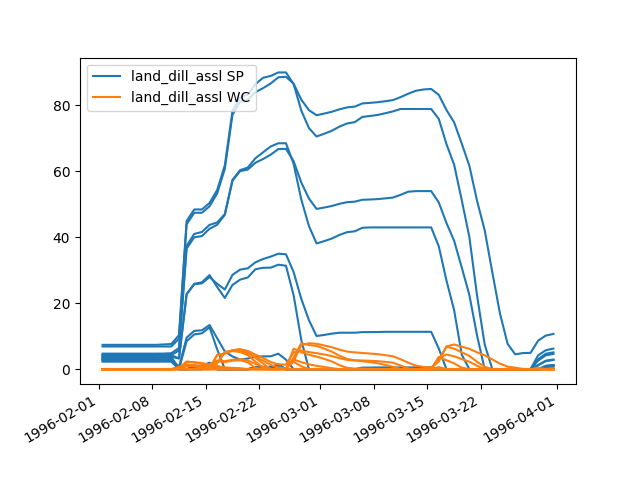
Alternatively, you can print the averaged time series by assigning
Trueto the argument average. We demonstrate this functionality for the factor sequenceTC(this time, without focusing on the time-series y-extent):>>> figure = land.plot_factorseries("tc", colors=("grey",)) >>> figure = land.plot_factorseries( ... "tc", average=True, focus=False, colors="black", linewidths=3) >>> save_autofig("Element_plot_factorseries.png", figure)
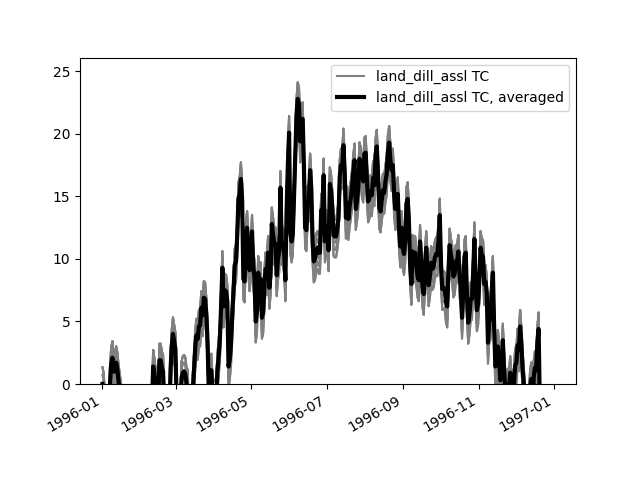
- plot_factorseries(*sequences: IOSequence | type[IOSequence] | str, average: bool = False, labels: tuple[str, ...] | None = None, colors: str | tuple[str, ...] | None = None, linestyles: Literal['-', '--', '-.', ':', 'solid', 'dashed', 'dashdot', 'dotted'] | tuple[Literal['-', '--', '-.', ':', 'solid', 'dashed', 'dashdot', 'dotted'], ...] | None = None, linewidths: int | tuple[int, ...] | None = None, focus: bool = True) Figure[source]¶
Plot the factor series of the handled model.
See the documentation on method
plot_inputseries()for additional information.
- plot_fluxseries(*sequences: IOSequence | type[IOSequence] | str, average: bool = False, labels: tuple[str, ...] | None = None, colors: str | tuple[str, ...] | None = None, linestyles: Literal['-', '--', '-.', ':', 'solid', 'dashed', 'dashdot', 'dotted'] | tuple[Literal['-', '--', '-.', ':', 'solid', 'dashed', 'dashdot', 'dotted'], ...] | None = None, linewidths: int | tuple[int, ...] | None = None, focus: bool = True) Figure[source]¶
Plot the flux series of the handled model.
See the documentation on method
plot_inputseries()for additional information.
- plot_stateseries(*sequences: IOSequence | type[IOSequence] | str, average: bool = False, labels: tuple[str, ...] | None = None, colors: str | tuple[str, ...] | None = None, linestyles: Literal['-', '--', '-.', ':', 'solid', 'dashed', 'dashdot', 'dotted'] | tuple[Literal['-', '--', '-.', ':', 'solid', 'dashed', 'dashdot', 'dotted'], ...] | None = None, linewidths: int | tuple[int, ...] | None = None, focus: bool = True) Figure[source]¶
Plot the state series of the handled model.
See the documentation on method
plot_inputseries()for additional information.
- Handles a
- hydpy.core.devicetools.clear_registries_temporarily() Generator[None, None, None][source]¶
Context manager for clearing the current
Node,Element, andFusedVariableregistries.Function
clear_registries_temporarily()is only available for testing purposes.These are the relevant registries for the currently initialised
Node,Element, andFusedVariableobjects:>>> from hydpy.core import devicetools >>> registries = (devicetools._id2devices, ... devicetools._registry[devicetools.Node], ... devicetools._registry[devicetools.Element], ... devicetools._selection[devicetools.Node], ... devicetools._selection[devicetools.Element], ... devicetools._registry_fusedvariable)
We first clear them and, just for testing, insert some numbers:
>>> for idx, registry in enumerate(registries): ... registry.clear() ... registry[idx] = idx+1
Within the with block, all registries are empty:
>>> with devicetools.clear_registries_temporarily(): ... for registry in registries: ... print(registry) {} {} {} {} {} {}
Before leaving the with block, the
clear_registries_temporarily()method restores the contents of each dictionary:>>> for registry in registries: ... print(registry) ... registry.clear() {0: 1} {1: 2} {2: 3} {3: 4} {4: 5} {5: 6}
